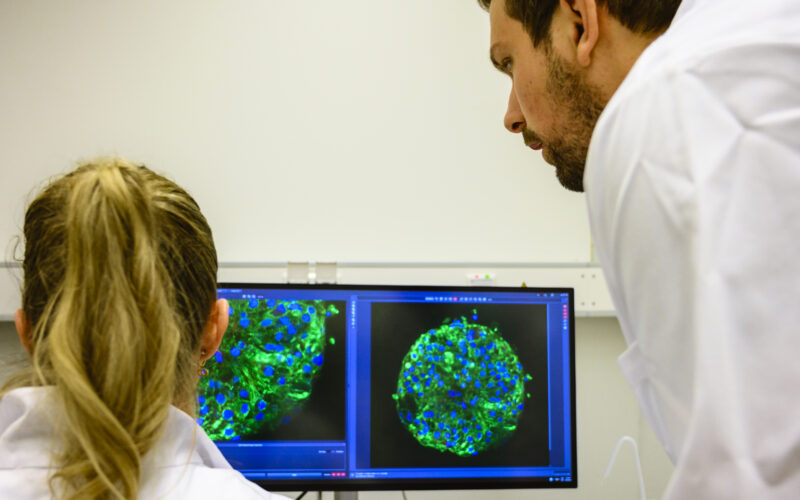
To better understand the human body, its diseases and to test new treatments, researchers use representative models. These include everything from simple cellular models for studying cell behaviour or reactions to a drug to big laboratory animal models to test the long-term effects of a disease or treatment on a body.
Until recently, using laboratory animals was the only way to model the complexity of the human body. However, while animals provide a good model for different organ systems and their interactions, the difference in species makes translation difficult. Researchers at the UMC Utrecht are developing various human cell models to better understand human-specific mechanisms and reduce the use of laboratory animals in research.
Cell models to find early biomarkers
Depending on the disease and its mechanism in the body, a two or three-dimensional cell culture can gain insight in the affected processes and how they might be diagnosed or treated. For cardiac amyloidosis, which is being studied by Alain van Mil at the UMC Utrecht Experimental Cardiology group, a human cell model was the most suitable. Van Mil developed the cell model to pursue an early diagnosis for transthyretin amyloid cardiomyopathy (ATTR-CM), one of the most common types of cardiac amyloidosis. This project is a translational research line, bridging Experimental Cardiology with the National Amyloidosis Expert Center for ATTR cardiac amyloidosis led by cardiologist Marish Oerlemans.
Cardiac amyloidosis is a build-up of fibrils in the heart muscle
ATTR-CM is characterized by the accumulation of fibrils in the heart muscle, causing cardiac dysfunction with dismal prognosis. The disease can be caused by many different genetic mutations in the gene encoding transthyretin (TTR). Such mutations lead to the production of a mutated unstable TTR protein in the liver, the primary organ that produces TTR protein. ‘The functional protein is made up of four subunits arranged like a four-leaf clover,’ Van Mil explains, ‘but in ATTR-CM it breaks off into its individual subunits. These then misfold and aggregate into so-called fibrils, which get distributed all over the body through the bloodstream and accumulate in the heart muscles.’
“The functional protein is made up of four subunits arranged like a four-leaf clover, but in ATTR-CM it breaks off into its individual subunits. These then misfold and aggregate into so-called fibrils, which get distributed all over the body through the bloodstream and accumulate in the heart muscles.”
A genetic and age-related disease
While the genetic mutations causing ATTR-CM are rare, the disease can also manifest, independent from mutations, as patients age, characterising the wild-type variant, which is much more common. ‘Just like in the genetic disease, transthyretin becomes unstable and breaks into monomers, which misfold, aggregate and form fibrils that accumulate in the heart. The reason for this spontaneous instability of the protein has not yet been found.’
In both variants, the proteins form fibrils that sit between heart cells and restrict the contraction of the heart muscle. Over time, fibrils accumulate, making the heart muscle stiffer and ultimately causing heart failure. Unfortunately, as this happens in a slow progression, and symptoms vary, patients are often diagnosed when the disease is already advanced to a late stage.
Currently, ATTR-CM can be halted but not cured
Only when caught early, ATTR-CM can be treated by slowing its progression. There are novel drugs that can stabilize the tetramer in the blood, decreasing the number of fibrils, and gene silencing using siRNA’s can “knock out” the mutant RNA in the liver cells. Another treatment option for the genetic variants is liver transplantation, which replaces the mutant-producing liver cells. However, all these therapies can only slow down the disease and not reverse the damage that has already been done. Patients are often already dealing with heart failure when the disease is diagnosed, and once heart failure develops overall survival is poor.
An early diagnosis is essential for treatment
To be able to treat patients as effectively as possible, the diagnosis needs to be as early in the disease as possible. Van Mil: ‘That’s why we are working on finding an early indicator for the disease to identify patients as early as possible. The ideal marker should be detectable in the blood during the early stages of the disease, allowing for monitoring during annual check-ups.’
“We’re working on finding an early indicator for the disease to identify patients as early as possible. The ideal marker should be detectable in the blood during the early stages of the disease, allowing for monitoring during annual check-ups.’
Creating the right disease model
To find this marker, a representative disease model is required. The researchers first employed 2D human-induced pluripotent stem cell-derived cell models using three major cell types present in the heart – cardiomyocytes, endothelial cells and fibroblasts. The models allow the group to focus on the details. ‘We do in vitro studies to look at which specific cell functions are affected by the disease,’ Van Mil says. ‘What we found is that when we add fibrils to the different cell types, we can soon see the function of the cells deteriorating. For the cardiomyocytes, the fibrils cause a decrease in cell viability and disrupt contractility, metabolism and calcium handling.’
Adding complexity as needed
By focusing on the details of these early cardiac cellular disease phenotypes, the group hopes to find an early biomarker unique to the disease. ‘We started simple, with 2D culture, but now we have moved on to 3D multi-cell type organoid-like tissues such as cardiac spheroids, but also bigger engineered cardiac tissues. These better recapitulate the complexity of the adult heart tissue, making them more predictive models’ Van Mil explains. ‘The next step is to see if we can model the disease in a flow system on a chip. This will allow us to mimic the disease even better, implement easy and relevant functional readouts, next to sampling for biomarker discovery’.
“We started with 2D culture, but we have moved on to 3D multi-cell type organoid-like tissues. The next step is to see if we can model the disease in a flow system on a chip. This will allow us to mimic the disease even better.”
The rise of organ-on-a-chip models
In the early 2000s, researchers in Utrecht discovered that adult-specific stem cells could organize themselves into miniature versions of organs when cultured in three-dimensional conditions. These now-called organoids show some of the same tissue structure and functions as the organ they are derived from, opening new possibilities for research. To better simulate the dynamic nature of the human body, researchers developed a culturing system connected by microfluidic channels. The resemblance of these systems to computer chips gave rise to the name: organ-on-a-chip or OOC model.

A new layer of complexity in models
Organ-on-a-chip models are more complex than simple, static cell models yet less complex than animal models. This allows for insight into specific processes and interactions while controlling or manipulating the experimental conditions. The models are currently used in fundamental research, diagnostics, drug testing, and a variety of other applications.
For example, organ-on-a-chip models can be used to test therapies for patients with complicated diseases that require a personalized treatment plan. If the organoid cultured with the patient’s stem cells responds well to the treatment, chances are, so will the patient.
However, the human body consists of more than one organ. To make the models more realistic, researchers have been working on combining multiple organoid types on one chip, creating a multiple organ-on-a-chip model (Multi-OOC). One of these researchers is Bas van Balkom, assistant professor at Regenerative Medicine Utrecht, UMC Utrecht.
“Each organoid has been developed in specialised culture conditions mimicking their original organ environment. Combining these features is challenging. We need circulation, a medium that keeps both organoids functional, all within the confines of one small chip.”
‘Fortunately,’ he says, ‘just like their counterparts in the human body, organoids seem to work with each other to maintain homeostasis, and show improved viability and function when cultured together. For example, a liver-organoid detoxifies the medium, while a lung organoid helps to add oxygen to the system.’
Using OOCs for the development of extracellular vesicle-based therapy
Bas van Balkom is working on developing a combined Kidney-Liver-Lung model to study the effects and distribution of vesicle-based therapies. Vesicles, or extracellular vesicles (EVs), have been an object of study since a puzzling discovery in mice.
When studying stem cell therapy to treat myocardial infarction in mice, researchers saw that the stem cells seemed to have a positive effect on heart function. Yet they could rediscover only three percent of the injected cells in the tissue. They concluded that the therapeutic effect must come from a secreted factor of the stem cells rather than the cells themselves.
Later, researchers at the Cardiology department of the UMC Utrecht discovered that the active component secreted by stem cells must be extracellular vesicles. As their name suggests, extracellular vesicles are cell-membrane-bound structures released into the bloodstream. They can vary in size and composition but usually contain RNA, proteins, and other molecules.
Since this discovery, researchers have been trying to understand why the EVs isolated from (mesenchymal) stem cell cultures have therapeutic effects. If these effects can be harnessed, a treatment with EVs could offer a cell-free, off-the-shelf therapeutic option for several diseases. In contrast to other human-derived treatments, it does not trigger an immune response and may even suppress it.
A human model to study the effects of EVs
EVs tend to find their way to injured organs, perhaps because the damage causes leaky vessels, or because local changes make these place more ‘sticky’ for EVs – or both. ‘We saw the same thing happening in the human kidney-liver-multi-OOC model that we developed,’ Van Balkom says. ‘Vesicles accumulated in the injured kidney organoid, recovering the barrier and transport function.’
“We found that the liver organoids, which were never exposed to injury, were also affected by the oxidative stress injury of the kidney, a very clear demonstration that our model mimic organ-organ interactions just like in living beings.”
‘Even more interesting,’ he continues, ‘is that we found that the liver organoids, which were never exposed to injury, were also affected by the oxidative stress injury of the kidney, a very clear demonstration that our model mimic organ-organ interactions just like in living beings’ Van Balkom says. ‘As vesicles stick to injured organs, they accumulated in both the liver and kidney organoids when injected into the medium. While this was never mentioned in the literature covering the animal experiments, we were able to identify the same effect when looking back at the data.’

Making the model even more human
Currently, Van Balkom’s group is trying to add more organs and tissues to the model to increase its complexity. ‘As vesicles also accumulated in the lung in other experiments, the next step is the addition of a lung organoid to the model. We also aim to add fat tissue to better model human disease and investigate how obesity influences systemic disease development and the effects of drugs.’
Towards a human-on-a-chip?
The UMC Utrecht is at the forefront of developing better models for human disease. In Regenerative Medicine Utrecht, groups from the UMC Utrecht, Utrecht University and the Hubrecht Institute collaborate every day by combining possibilities and sharing technologies.
The combinations of organs they are exploring could be expanded further and further, as research is moving towards a more dynamic and complex model of the human organ systems. The goal is to predict and model treatment outcomes for individual patients to optimise and personalise therapies. Ultimately organ-on-a-chip and eventually human-on-a-chip models may fully replace animal experiments in research and provide us with patient specific data.