PhD research by Vincent de Maat at UMC Utrecht has revealed new genetic techniques to study the genome of E. faecium and to more easily manipulate its genome to get a better understanding of its gene function. The results show that not a singular gene is responsible for its improved adaptation and survival allowing it to become a successful hospital associated disease, but a complex set of genes which are largely present across E. faecium strains.
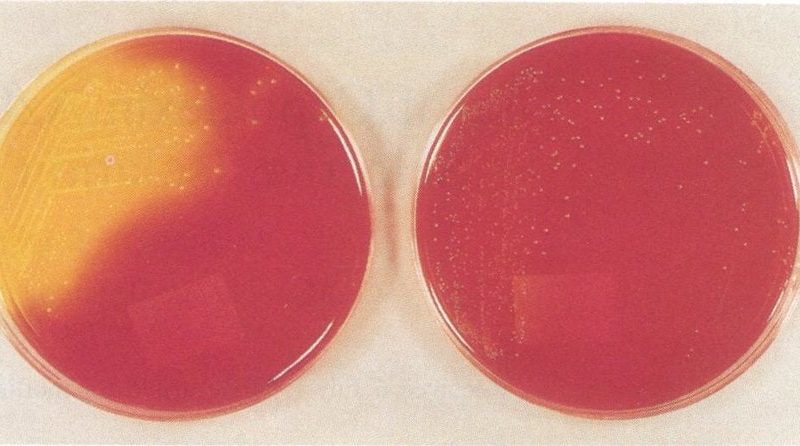
Bacteria live all around us and in general they live peacefully in balance with humans. Sometimes, however, this balance is disrupted when a bacteria starts to adapt and explore new areas. This is also the case for the Gram-positive gut bacterium Enterococcus faecium. Over the years, this bacterium has acquired many antibiotic resistances (e.g. to ampicillins, aminoglycosides, fluoroquinolones and vancomycin) and is increasingly causing hospital associated infections. The goal of the research by PhD candidate Vincent de Maat (Department of medical Microbiology, UMC Utrecht) was to unravel which adaptions E. faecium has made that allowed it to survive in blood or serum and to become resistant to cleaning alcohol, thereby becoming successful as a hospital associated disease.
For this research, Vincent and co-workers developed several new genetic techniques to study the genome of E. faecium and to more easily manipulate its DNA to get a better understanding of the function of the genes. The results show that not a singular gene is responsible for the improved adaptation, but a complex set of genes which are largely present across E. faecium strains. This indicates that this bacterium as a species is already highly suited to survive in hospitals. Further research needs to be conducted to understand in greater detail how this bacterium survives and discover new methods to prevent it from causing infections in hospitalized patients.
The research described in the thesis by Vincent de Maat has identified essential E. faecium genes that are important for its enhanced survival in hospitals, allowing it to more easily spread and subsequently colonizing more patients. In addition, new genes were identified that allow E. faecium to better survive in blood or serum as well as to becoming resistant to 2-propanol (cleaning alcohol). Many of these genes are present in most strains tested which were isolated from healthy humans, animals and patients, indicating that E. faecium is already naturally extremely adaptable to survive in hospitals. For this research, powerful genetic techniques were introduced that can both broaden and deepen insights into the genetics of E. faecium. This will eventually lead to a better understanding of the internal workings of E. faecium and provide new leads for preventative interventions or improved treatment modalities.
A hospital-acquired infection (HAI) is an infection that is acquired in a hospital or other health care facility (nursing home, rehabilitation facility, outpatient clinic, diagnostic laboratory or other clinical settings). Infection is spread to the susceptible patient in the clinical setting by various means, including health care staff, contaminated equipment, bed linens or air droplets. The infection can originate from the outside environment, another infected patient, or staff that may be infected, or in some cases, the source of the infection cannot be determined. In some cases the microorganism originates from the patient’s own skin microbiota, becoming opportunistic after surgery or other procedures that compromise the protective skin barrier. HAI can cause severe pneumonia and infections of the urinary tract, bloodstream and other parts of the body. Many types display antimicrobial resistance, which can complicate effective treatment.
Vincent de Maat (1988, Apeldoorn) defended his PhD thesis on February 24, 2022 at Utrecht University. The title of his thesis is “Adaptations of hospital-acquired Enterococcus faecium”. Supervisors were prof. dr. Rob Willems (Department of Medical Microbiology, UMC Utrecht) and prof. dr. Willem van Schaik (University of Birmingham, Birmingham, United Kingdom). Vincent works as a R&D scientist at Kinetic Evaluation Instruments in Leusden (the Netherlands), where he is involved in the development of new assays for the detection of Mycobacterium tuberculosis in human blood.