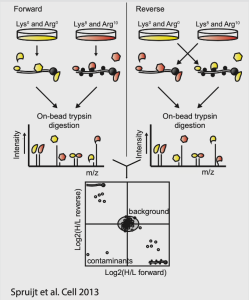
Experimental workflows
We provide the following experimental workflows:
- (Phospho)proteomics (with relative or absolute quantification).
- Protein affinity purification using epitope tags such as GFP, HA, FLAG or Myc tag.
- Peptide or DNA affinity purification (identification of proteins interacting with a specific (modified) peptide or DNA sequence of interest).
- PAQMAN for the determination of apparent Kd of transcription factors for a specific sequence.